Versatile antiviral proteins discovered with supercomputer
A single tiny molecule that can destroy flu, corona, HIV and Zika viruses? Yes, it really does exist. An algorithm that mimics evolution has produced protein molecules that work against Zika and HIV in the laboratory. Biophysicist Niek van Hilten, who received his doctorate with honors on 14 September, contributed to this discovery.
Virus vaccines and virus inhibitors face two problems: a specific drug is needed to combat each virus, and it only works temporarily because viruses evolve rapidly. Almost all drugs target proteins that stick out of the virus like flags. Niek van Hilten: ‘This has become an established practice over the years; almost all drugs do that. But every virus has unique proteins, and they are constantly changing.’
Molecule that senses curvature
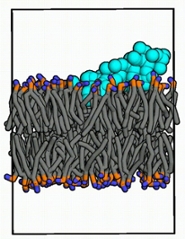
Biophysicist Van Hilten has been involved in research into antiviral drugs using a very different method of attack. ‘Many viruses are shaped like small spheres. Corona, influenza, HIV and Zika, for example, are enveloped viruses. They are almost a thousand times smaller than our own cells.’ They have a membrane similar to human cells: two layers of lipid molecules with the long tails facing each other (see image). ‘Because viruses are so small, their membrane is much more curved than that of our body cells. Some small proteins can sense this curvature and then puncture the membrane, destroying the virus.’
The existence of proteins that can fight these spherical viruses was first discovered 20 years ago. ‘They found a protein molecule called AH that recognises enveloped viruses by their membrane and then perforates it. Fatal for the virus.’ Researchers have already tested several variants on mice and macaques.
Designed from scratch for an accurate picture
A revolutionary antiviral drug may seem within reach once research has progressed to the point of testing on monkeys. But experience shows that a lot can go wrong on the long road to getting a drug to market, says Van Hilten. This is why his and his colleagues’ research is so important. ‘We use the AH protein as an example to understand exactly how such a drug works and to design such a protein from scratch.’ And it worked.
The protein molecules that recognise viral membranes consist of 24 blocks of one amino acid. Van Hilten: ‘There are 20 different amino acids, which means there are 2024 ways to build a protein from 24 blocks. That’s about 17 followed by 30 zeros.’ Too many to test virtually, even for an algorithm capable of supercomputing.
Protein evolution thanks to an algorithm
Fellow PhD candidate Jeroen Methorst therefore wrote an evolutionary algorithm that started with 144 random possible proteins. ‘This was the most efficient number that our supercomputers could handle.’ In a virtual piece of membrane developed by Van Hilten, the algorithm examined how well these proteins could identify and perforate a curved viral membrane. It then allowed the best ones to reproduce with each other. ‘After 25 generations, the molecules’ performance was no longer improving, so we stopped.’
In Leiden and in the German city of Ulm, fellow researchers synthesised several promising variants. ‘In laboratory tests, some of these proteins proved effective against Zika and HIV and were safe for human cell membranes. When I heard that, it was the best moment of my PhD.’
Film of molecules helps researchers
An innovative feature of the algorithm used by the researchers is the combination of an evolutionary algorithm with molecular dynamics simulations. Van Hilten: ‘The latter is a technique for simulating the movements and interactions of molecules. This results in a video (see video below, Ed.) that shows what the molecules are doing. Many other algorithms work like a black box and churn out a result; our algorithm allowed us to see what happened for each result.’ Van Hilten was able to integrate molecular dynamics into the algorithm. ‘This allows us to ask algorithms like this all sorts of other development questions, where you can then see what the resulting molecules do.’
The movements and interactions of molecules
Due to the selected cookie settings, we cannot show this video here.
Watch the video on the original website orAs he prepares to take up a postdoc position in San Francisco, Van Hilten leaves his colleagues with wonderful new possibilities, but also with new questions. ‘Why didn’t all the molecules suggested by the algorithm work? Probably because our model was still very simple. For example, the algorithm only looked at the behaviour of proteins in very simple membranes made of one type of lipid molecule. In biological reality, there are up to 50 different types.’
Text: Rianne Lindhout
