Caught in living cells: how bacteria regulate their genes to defend themselves
For the first time, it was shown in living cells how the bacterium E. coli regulates genes that help it survive in a new environment. Biochemist Fatema Zahra Rashid managed to do this using a technique she fine-tuned. Her research into changes in 3-dimensional chromosome structure offers clues for ways to fight pathogens and appeared in Nature Communications on November 17.
Bacteria are a common threat to human health and well-being. Whereas some are known as causing serious food poisoning, others can cause life-threatening infections of tissues, such as human lungs. In all cases, to be effective at their ‘task’, the bacteria need mechanisms to survive by adapting to their environment.
The bacterium and its chromosome
You may consider bacteria to be little bastards. But it is really fascinating how single-celled organisms, often considered simple, are able to adapt so well to a hostile environment. If we find out how, we might be able to stop them from doing so and thus fight off pathogens.
The chromosome of bacteria floats freely in the cell, but is strongly compacted in order to fit. If you could zoom in on a part of the chromosome, you would see that the DNA is tightly or loosely folded, with bends, and loops here and there that connect a piece of DNA to a piece of DNA further away. All kinds of proteins take care of that. By being bound or unbound, they regulate which genes are accessible and can be read and thus serve as templates for making proteins.
A group of genes
For decades, biochemists and geneticists have been studying how the regulation of genes works. Fatema Zahra Rashid has been one of them for about ten years: first as a master's student, then as a PhD student and now as a postdoc. 'My research focuses on a specific piece of DNA in E. coli: the ProVWX or ProU operon. That is a group of genes that we know are involved in protecting against an osmotic shock.' An osmotic shock occurs when the environment is suddenly much saltier or less salty than the fluid inside the bacteria. The amount of salt then also changes inside and this leads to cellular processes malfunctioning, and cells dying, if they are not able to adapt.
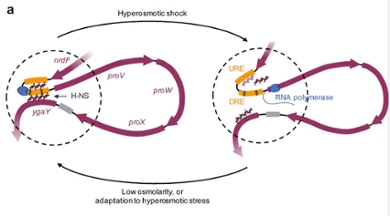
A protein binds DNA pieces together with two hands
A protein binds DNA pieces together with two hands
With such an osmotic shock, the DNA in the aforementioned ProU operon is suddenly read heavily. In test tubes with pieces of DNA, this had already been shown to be related to the regulatory protein H-NS, found in many bacteria. Rashid's group leader, Professor of Molecular and Cellular Biochemistry Remus Dame, showed as a PhD student how this protein grabs two parts of a DNA with two ‘hands’ and makes a loop, he says. 'Upon osmotic shock, we thought, those little hands could release making the DNA available for reading and making a specific protein system protecting the cell. When equilibrium is restored in the cell, the little hands would grab the DNA again and transcription stops.’
A snapshot of the cell
Dame’s colleague Rashid has now shown in living bacteria that this is indeed how it works. She exposed living bacteria to an osmotic shock. 'After a while, I added a fixative, causing all the proteins and DNA to ‘freeze’. In a way, this is taking a snapshot of the cell.' From those fixed cells, Rashid extracted the DNA and cut it into small pieces. If the operon was in a loop, the start and end pieces were stuck together. If not, the start and end pieces were further apart. She could measure that. She was also able to determine that more of the protection system was produced upon osmotic shock based on the genes from ProU. She normally speaks to her supervisor Remus Dame once a week. 'When I first saw these results about two years ago, I didn't wait for my weekly consultation,' she smiles.
Acid or hot
A remedy to tackle pathogens based on their DNA regulation is not there anytime soon. A milestone is nonetheless there. Dame: 'These kinds of regulation mechanisms have been studied for much longer in eukaryotes, organisms with a cell nucleus. We now see that they are also highly developed in prokaryotes. Several PhD students are now using this method to see whether the same thing happens in bacteria when the environment suddenly becomes very acidic, as in the stomach. And what happens when there is a change in temperature, such as when a bacterium moves from environment to host.'
Text: Rianne Lindhout
